Prostate Cancer
As early as in September 2010, NGS technologies was still in its infancy with barely a few datasets from cancer samples in the public repository useful in training. IBAB found the only RNA-seq dataset from prostate cancer from 3 disease individuals with matched-normal
samples for use as control. This dataset was used to train PGDB-2011 and 2012 batches in NGS data analysis. Tasks included extraction of various differential genetic elements including cancer-specific gene/non-coding/splice variant expression and SNPs. Considering that
tools/pipelines for analysis were still unavailable, students had to develop methods and write programs to extract biologically meaningful entities from the RNA-seq dataset. We dared publish our results providing us with much confidence about our training process required to
produce national and international level capacity in NGS data analysis.
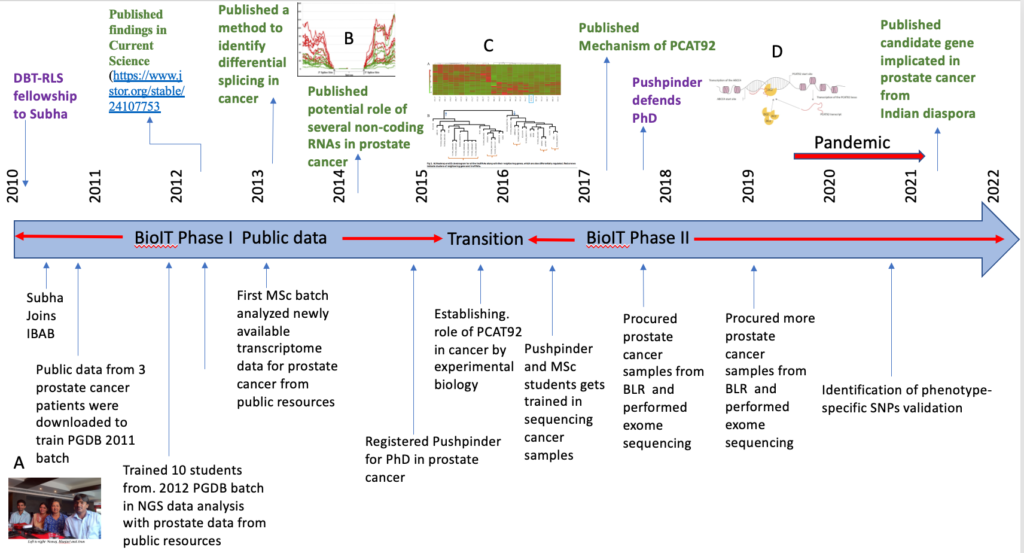
In 2013, as the public repository was growing, results from the previous dataset were validated with results from other datasets deposited by other investigators pertaining to prostate cancer. Our first effort was to identify pairs of differentially coregulated genes and non-coding genes from the same locus, in prostate cancer. One of the embedded figure C in the timeline above shows such pairs (PMID: 25933431), which were both up- and down-regulated. Since Subha was a PI at NIH, she had also secured access to a large control dataset from prostate cancer, which was used to validate these findings. One of the pairs mentioned above had a gene that played a role in androgen transport (ABCC4) and the corresponding non-coding gene (PCAT92) from the same locus was already known to be associated with prostate cancer. The hypothesis was that PCAT92 may play a role in prostate cancer by regulating ABCC4 expression. Deciphering the mechanism of ABCC4 expression regulated by PCAT92 was to become part of a PhD thesis. Figure D in the timeline above shows that PCAT92 recruits ZIC2, a transcription factor, to the site by simultaneously binding to the both the chromosomal DNA near ABCC4 promoter site and PCAT92 to aid ABCC4 expression (PMID: 3019775).
The functional validation of our analysis using the public data during the first phase of BioIT together with the merger of BioIT with IBAB gave us a boost during the phase of BioIT to dare attempt identification of candidate genes in prostate cancer in the context of Indian diaspora. We procured samples from 58 prostate cancer patients from Dr. Raghunath (Table 1 above) and generated 390 Gb of exome sequencing data, which is currently available under the accession PRJNA838939. The clinical parameter as shown in the table allowed for in-depth context-specific analysis and interpretation of data. For example, we found that AR gene is most highly mutated in the T3 tumors with lymph-node not involved subset. Patients at T3 stage have the highest number of CNVs. Furthermore, POLQ, a novel target gene is mutated at a frequency of 53.8% in recurrent with only 20% in no-recurrent cases. Findings from this work is now published in peer reviewed journal (PMID: 35737091).